|
IMPUTE version 2 (also known simply as IMPUTE2) is a genotype imputation program
based on ideas from
Howie et al. (2009).
Please click on the links below to download the software or learn how to use it.
Page last updated Aug 5, 2010. We will be adding a number of features to the website in the near future, so please check back if you are actively using the software. New data! We have now posted the latest haplotypes from the 1,000 Genomes Project, which were released in June 2010. There are 120 CEU haplotypes, 120 CHB+JPT haplotypes, and 118 YRI haplotypes in the new dataset. You can download the official release haplotypes or a set of haplotypes tailored to work with HapMap 3 below; we recommend using the latter (1,000 Genomes + HapMap 3) reference set for most imputation tasks. New strategies! We have been working hard to show how IMPUTE2 can use large reference panels with diverse ancestry to improve the imputation of rare alleles and eliminate the need to choose which haplotypes will form the reference set. You may have seen us talk about this work; it is not yet published, but we have written a short summary of our ideas, results, and motivations here. We have also used these ideas to inform the packaging of the 1,000 Genomes and HapMap 3 reference sets; you can download haplotypes that fit IMPUTE2's reference panel philosophy here. IMPORTANT: The population-genetic approximation used by IMPUTE2 is only valid over short genomic distances -- if you use the program on too large a region, the quality of the inference will diminish. While the definition of "short" depends on various characteristics of a dataset, in general the program should only be used on regions of 5 Mb or shorter. If you need to impute a longer region (e.g., a whole chromosome in a genome-wide association study), we provide simple command-line options for splitting chromosomes into smaller chunks for analysis: this process in described in the section on Analyzing Whole Chromosomes. |
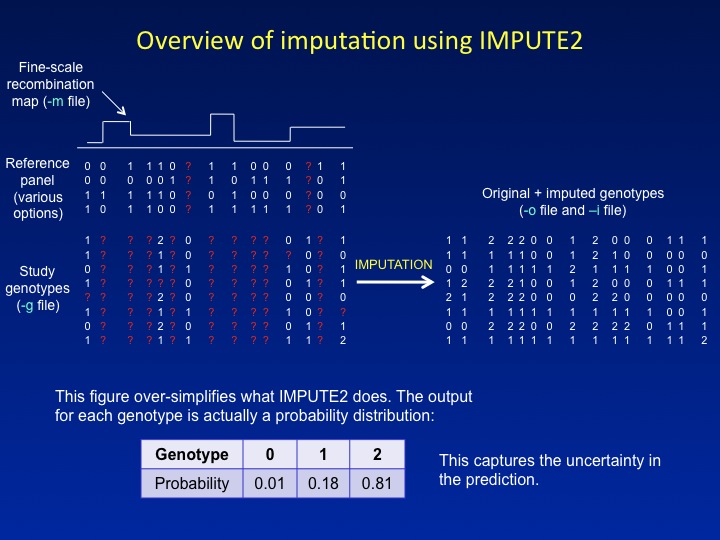
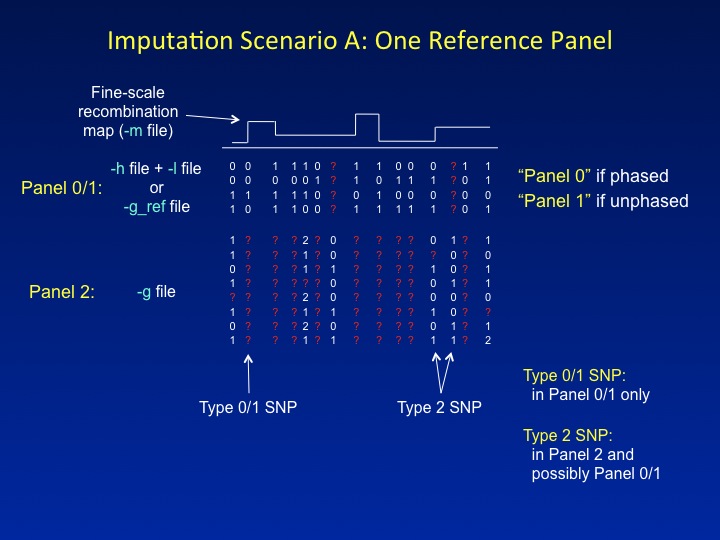
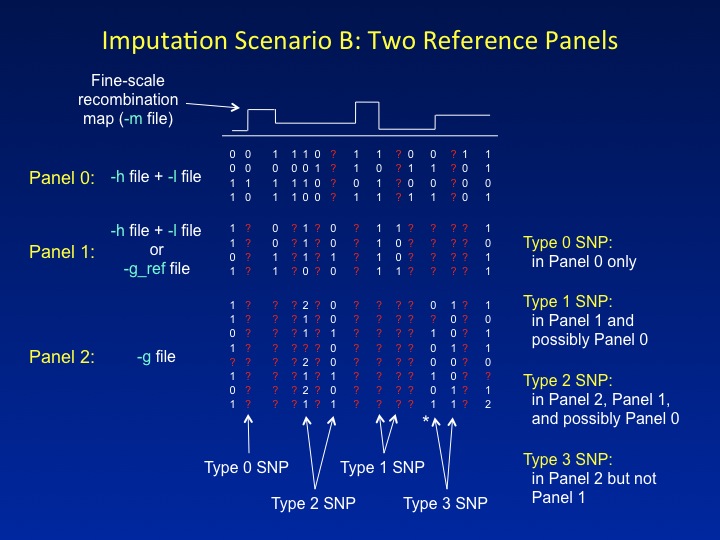
|
Platform
|
File
|
|
Linux (x86_64) Static Executable
|
impute_v2.1.0_x86_64_static.tgz
|
|
Mac OS X Intel
|
impute_v2.1.0_MacOSX_Intel.tgz |
| tar -zxvf impute_v2.X.Y_i386.tgz |
|
Haplotype, legend, sample, and genetic map files These downloads contain the data needed to impute genotypes using reference panels from HapMap 3 and the 1,000 Genomes Project. Each dataset includes the latest haplotypes from the 1,000 Genomes panel of interest, along with all available HapMap 3 haplotypes, except those present in the relevant 1,000 Genomes panel. We remove these duplicate haplotypes so that the two datasets can be combined without causing "double counting" of haplotypes during imputation. Both sets of haplotypes have also been filtered to remove SNPs with apparent quality issues. To see an example command that combines HapMap 3 and 1,000 Genomes haplotypes in a single imputation analysis, go here. To see our rationale for using all HapMap 3 haplotypes together, rather than focusing on population-matched subsets, go here. To learn more about our scheme for filtering out low-quality SNPs, go here. If you prefer unfiltered 1,000 Genomes haplotypes, you can download them from here; similarly, you can download unfiltered HapMap 3 haplotypes from here. |
Download packages (warning: large files) [CEU] [YRI] [CHB+JPT (coming soon)] |
|
Haplotype, legend, sample, and genetic map files These downloads contain the data needed to impute genotypes using reference panels from the 1,000 Genomes Project. The files are unfiltered, in the sense that we have not modified them from the official release versions. These haplotypes are generally of high quality, but they may contain a small fraction of poorly genotyped SNPs. |
Download packages (warning: large files) [CEU] [YRI] [CHB+JPT (coming soon)] |
|
Haplotype, legend, sample, and genetic map files These downloads contain the data needed to impute genotypes using reference panels from HapMap Phase 3. The files are unfiltered, in the sense that we have only modified them minimally from the official release versions. These haplotypes are generally of high quality, but they may contain a small fraction of poorly genotyped SNPs. In HapMap 3, the most common problem is that an allele will "drop out" of the genotyping assay, thereby making every individual appear homozygous for the same allele. |
Download packages (warning: large files) [ALL PANELS] |
|
./impute2 \ |
|
./impute2 \ |
|
./impute2 \ |
|
./impute2 \ |
|
./impute2 \ |
|
./impute2 \ |
|
./impute2 \ |
| Flag | Default | Description |
|
-g <file>
REQUIRED |
none | File containing genotypes for a study cohort in which we want to impute untyped SNPs. The format of this file is described on our file format webpage and is the same as the output format from our genotype calling program CHIAMO. |
|
-m <file>
REQUIRED |
none | Fine-scale recombination map for the region to be analyzed. This file should have three columns: physical position (in base pairs), recombination rate between current position and next position in map (in cM/Mb), and genetic map position (in cM). The file should also have a header line with an unbroken character string for each column (e.g., "position COMBINED_rate(cM/Mb) Genetic_Map(cM)"). |
| -h <file 1> <file 2> | none | File of known haplotypes, with one row per SNP and one column per haplotype. In IMPUTE2, it is possible to specify two known haplotypes files. In this case, the file with more SNPs should be provided first (in the <file 1> position) and the file with fewer SNPs should be provided second (in the <file 2> position), with a single space separating the file names. Every haplotype file needs a corresponding legend file (see below), and all alleles must be coded as 0 or 1 -- no other values are allowed. |
| -l <file 1> <file 2> | none | Legend file(s) with information about the SNPs in the -h file(s). Each file should have four columns: rsID, physical position (in base pairs), allele 0, and allele 1. The last two columns specify the alleles underlying the 0/1 coding in the corresponding -h file; these alleles can take values in {A,C,G,T}. Each legend file should also have a header line with an unbroken character string for each column (e.g., "rsID position a0 a1"). When using two known haplotypes files with IMPUTE2, you must supply the corresponding legend files in the same order -- i.e., the file with more SNPs comes first. |
| -g_ref <file> | none | File containing unphased genotypes to use as a reference panel for imputation. This file should follow the same format as the -g file. A -g_ref file can be used as the lone reference panel for imputation, or it can be combined with a single -h file to create a two-tiered reference panel (in the latter case, the -g_ref file should contain roughly a subset of the SNPs in the -h file). |
| Flag | Default | Description |
|
-int <lower> <upper>
REQUIRED |
none |
Genomic interval to use for inference, as specified by
<lower>
and
<upper>
boundaries in base pair position. The boundaries can be expressed either in long form (e.g.,
|
| -buffer <int> | 250 kb | Length of buffer region (in kb) to include on each side of the analysis interval specified by the -int flag. SNPs in the buffer regions inform the inference but do not appear in output files. Using a buffer region helps prevent imputation quality from deteriorating near the edges of the analysis interval. |
| -Ne <int> | 14000 |
"Effective size" of the population (commonly denoted as Ne in the population genetics literature)
from which your dataset was sampled. This parameter scales the recombination rates that IMPUTE2
uses to train its population model. As a starting point, we suggest values of
11418
for imputation from HapMap CEU,
17469
for YRI, and
14269
for CHB+JPT.
When combining reference panels, we suggest taking the average of the panel-specific Ne values, weighted by the number of chromosomes in each panel; e.g., for a CEU+YRI+CHB+JPT panel in HapMap Phase II data, the Ne would be |
| -call_thresh <float> | 0.9 |
Threshold for calling genotypes in the
-g file.
For each individual at each SNP, the program will use the genotype with the maximum probability
if that probability exceeds the threshold; otherwise, the genotype will be treated as missing.
NOTE: This threshold only applies to input genotypes. If you want to apply a calling threshold to IMPUTE2's output probabilities, you will have to do it yourself. However, it is usually not a good idea to treat imputation output this way; see the webpage of our association-testing software SNPTEST for better suggestions. |
| -nind <int> |
# of indiv in |
Number of individuals from the
-g file
to include in the analysis. For example, to impute only the first five individuals, set
|
| Flag | Default | Description |
| -strand_g <file> | none |
File showing the strand orientation of the SNP allele codings in the
-g file,
relative to a fixed reference point. Each SNP occupies one line, and the file should have
two columns: (i) the base pair position of the SNP, and (ii) the strand orientation ('+' or '-')
of the alleles in the genotype file; the columns should be separated by a single space.
The ordering of the SNPs in this file does not matter (by contrast to the
-g file,
which must be sorted by SNP position),
and it is okay if some SNPs in the strand file are not present in the genotype file
(e.g., due to filtering). Some model strand files are included in the
Example/
directory that comes with the software download.
NOTE: This flag replaces the -s flag from versions prior to v2.1.0. |
| -strand_g_ref <file> | none |
Same as
-strand_g,
but applies to the
-g_ref file.
NOTE: This flag replaces the -s_ref flag from versions prior to v2.1.0. |
| -fix_strand_g |
Activates the program's internal strand alignment procedure for the
-g file
(Panel 2).
The strand is aligned to the alleles in Panel 0, if present, otherwise to
Panel 1. The strand is aligned deterministically where possible (e.g.,
flipping A/C in Panel 2 to match G/T in the reference) and by allele frequency otherwise
(at A/T and C/G SNPs, whose alignment cannot be resolved by labels alone); in the latter
case, the program codes the alleles such that Panel 2 and the alignment
reference (Panel 0 or 1) have the same minor allele.
NOTE: This flag can be used in conjunction with the -strand_g option. In that case, the information from the strand file takes precedence, i.e., the program will not try to "fix" the strand of SNPs that have explicit strand info already. This is useful if you have strand information for some SNPs but not others. NOTE: You should take care when using this option. In particular, it can get the alignment wrong at A/T and C/G SNPs with minor allele frequencies near 50%, which can hurt the inference by distorting the local haplotype patterns. The only way to get the correct alignment at these kinds of SNPs is to track down the original assay and determine which strand was measured. NOTE: This flag replaces the -fix_strand flag from versions prior to v2.1.0. |
|
| -fix_strand_g_ref |
Similar to
-fix_strand_g,
but applies to the
-g_ref file
(Panel 1). In this case the strand is aligned to the alleles in
Panel 0, so the flag does not work if this panel is not present.
NOTE: Just as -fix_strand_g can be used in conjunction with -strand_g, this flag can be used in conjunction with the -strand_g_ref option. As before, the strand file takes precedence over the internal strand-fixing procedure. NOTE: As with -fix_strand_g, you should be careful about using this option to align A/T and C/G SNPs with minor allele frequencies near 50%. NOTE: This flag replaces the -fix_strand_ref flag from versions prior to v2.1.0. |
| Flag | Default | Description |
| -exclude_snps_g <file> | none |
List of SNPs to exclude from the
-g file.
The list should take the form of a single column of identifiers in a text file. The SNPs can be identified
by their SNP IDs (first column of
-g file),
their rsIDs (second column of
-g file),
or their base pair positions (third column of
-g file).
Excluded SNPs will be treated as if they had not been present in the genotypes file,
and they will not be shown in the output unless you use the
|
| -exclude_snps_g_ref <file> | none | Same as -exclude_snps_g, but applies to the -g_ref file. |
| -impute_excluded |
Specifies that SNPs excluded from the study dataset via the
|
|
| -include_snps <file> | none | List of reference-panel-only SNPs to impute. If you do not want the program to impute all of the Type 0 and Type 1 SNPs in the region you are analyzing, you can use this list to specify a subset of SNPs to impute; all other SNPs will be ignored unless they have data in the -g file. The list should take the form of a single column of identifiers in a text file. The SNPs can be identified by their SNP IDs (first column of -g_ref file), their rsIDs (second column of -g_ref file or first column of -l file), or their base pair positions (third column of -g_ref file or second column of -l file). This option does not have any effect on Type 2 and Type 3 SNPs. |
| -sample_g <file> | none |
File of sample IDs for the individuals in the
-g file;
should follow the format described
here.
Only the first three columns are necessary, and only the first two columns are used by
IMPUTE2 (i.e., the third column can have dummy values, and subsequent columns
do not affect the algorithm).
NOTE: Currently, the only reason to provide a sample file is if you want to exclude some individuals via the -exclude_samples_g option. |
| -sample_g_ref <file> | none | Same as -sample_g, but applies to the -g_ref file. |
| -exclude_samples_g <file> | none |
List of samples to exclude from the
-g file.
The list should take the form of a single column of identifiers in a text file. The samples can be identified
by the IDs in either of the first two columns of the
-sample_g file,
which is REQUIRED
if you want to use this option. Excluded samples will be treated as if they had not been present in the
genotypes file, and the program will re-print the original sample list, minus the excluded samples,
to a file named
" NOTE: Part of the IMPUTE2 algorithm involves pooling information across the individuals in your study dataset. Samples with systematically aberrant genotypes (due, e.g., to degraded assay DNA) can confuse this part of the model; you should take care to identify such samples ahead of time and exclude them either manually or with this option. |
| -exclude_samples_g_ref <file> | none |
Same as
-exclude_samples_g,
but applies to the
-g_ref file.
One difference is that the program will not print a filtered list of
-g_ref
samples like the one that gets printed with
|
| Flag | Default | Description |
| -o <file> | ./test.impute2 | Name of main output file. Follows the same format as the -g file. |
| -i <file> | [-o]_info |
Name of SNP-wise information file with one line per SNP and a single header line at the beginning;
versions of IMPUTE prior to v2.1.0 did not print the header.
This file always contains the following columns (header tags shown in parentheses):
1. SNP identifier from -g file (snp_id) 2. rsID (rs_id) 3. base pair position (position) 4. expected frequency of allele coded '1' in the -o file (exp_freq_a1) 5. measure of the observed statistical information associated with the allele frequency estimate (info) 6. average certainty of best-guess genotypes (certainty) 7. internal "type" assigned to SNP (type) -- column did not exist prior to v2.1.0 Depending on the command-line options invoked, there may also be columns labeled info_typeX, concord_typeX, and r2_typeX, where X takes values in {0,1,2}. For SNPs that have genotypes in the -g file, concord_typeX is the concordance between the input genotypes (after applying the |
| -r <file> | [-o]_summary | Name of file that records a summary of the screen output. |
| -w <file> | [-o]_warnings | Name of file that records warnings generated by IMPUTE2. |
| -os <int> <int> ... | 0 1 2 3 |
"Output SNPs": specifies the SNP types that will be printed to the output file (SNP labeling
is discussed in the Overview). By default, all imputed and genotyped
SNPs are included in the output, i.e.,
" |
| -o_gz | Specifies that the main output file should be compressed by the gzip utility; this also applies to some non-standard output files that can become large. | |
| -outdp <int> | 3 | Specifies the number of decimal places to use for reporting genotype probabilities in the main output file. |
| -no_snp_qc_info | Suppresses printing of info_typeX, concord_typeX, and r2_typeX columns in -i file. | |
| -no_sample_qc_info |
Suppresses printing of per-sample quality control metrics file. The default is to print a file named
" |
|
| -phase |
IMPUTE2 always implicitly phases the study dataset
( In addition to this "best-guess" haplotypes file, the program also prints the certainty that each successive pair of heterozygous SNPs is correctly phased. These certainties occur in a file named " As shown in the examples section, it is possible to use the |
|
| -pgs |
"Predict Genotyped SNPs": Tells the program to replace the input genotypes from the
|
|
| -pgs_miss |
Unlike
WARNING: This is an appealing option that promises to simply "fill in" sporadically missing genotypes in your input data. However, we think that following this procedure and then testing the SNPs for association could cause subtle problems. We are investigating these issues, but in the meantime we suggest that you only use this option with great caution; using it naively may lead to bad results, and you do so at your own risk. |
| Flag | Default | Description |
| -seed <int> | random | Initial seed for random number generator. The seed is set using the system clock unless it is manually overridden with this option. |
| -no_warn | Turns warnings off, so that the -w file does not get printed. | |
| -no_fill | Turns hole-filling off, so that SNPs included in the -g file but not in the lowest reference panel cannot contribute to the inference. | |
| -no_remove | Prevents the program from discarding SNPs whose alleles cannot be aligned across panels. |
| cat chr16_chunk1.impute2 chr16_chunk2.impute2 chr16_chunk3.impute2 > chr16_chunkAll.impute2 |